Schematic illustration of Brownian dynamics simulation
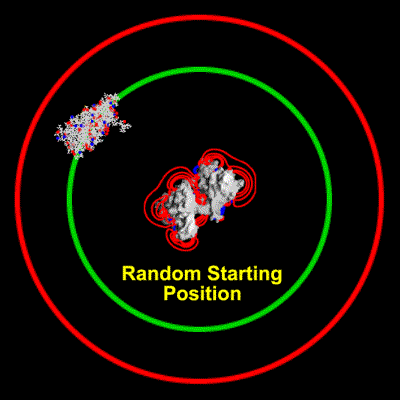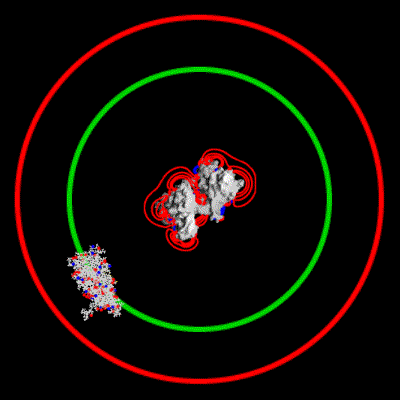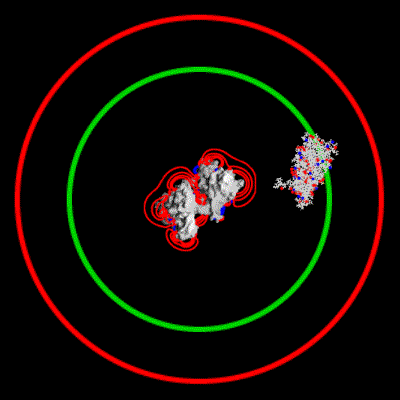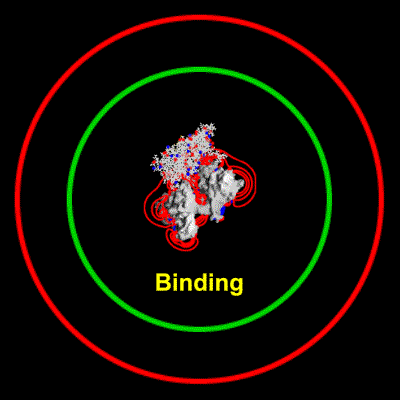

With advances in computational power in the past years, it has become possible to simulate the interaction of larger biomolecules, the polymerization of actin being one such system. Using Brownian dynamics simulations, the binding of a monomer to the end of a filament was simulated over a range of ionic strengths. The animation depicts two `fictional' trajectories intended to illustrate the basic concepts behind the simulations. For each trajectory, the monomer was started with a random orientation on a sphere surrounding the filament. By keeping track of the number of successful binding events at each end of the filament, a rate constant for binding can be calculated. This study showed that electrostatic interactions in fact lead to an asymmetry in polymerization rates between the two ends.
More details and analysis of the simulations will be published in:
D. Sept, A.H. Elcock and J.A. McCammon, Computer simulations of actin polymerization can explain the barbed-pointed end asymmetry, to appear in J. Mol. Biol., 1999or you can contact Dave Sept directly.